Author: Denis Avetisyan
Researchers have developed a powerful framework for accurately reconstructing complex network structures from observational data, even in directed graphs.
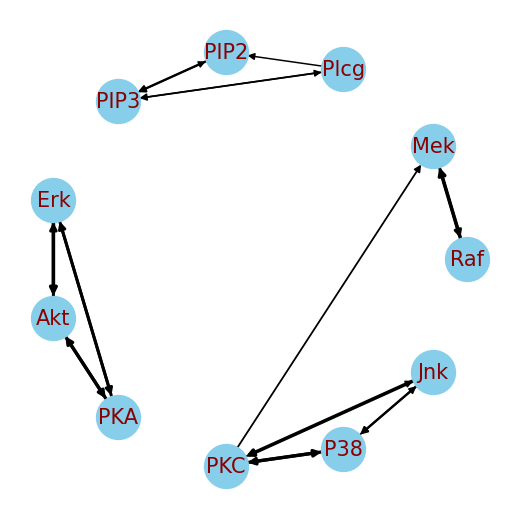
![The reconstructed graph stands as testament to the method detailed in reference [3], a lineage traced through careful derivation and iterative refinement.](https://arxiv.org/html/2601.15999v1/imgs/real_data/compare.png)
This review details a covariance matching method leveraging Riemannian optimization for improved graph topology identification and structural equation modeling.
Inferring the structure of networked systems from observational data remains a significant challenge, often hampered by the need for strong assumptions or computationally intractable optimization. This paper introduces a novel framework, ‘Graph Topology Identification Based on Covariance Matching’, which directly aligns empirical data covariance with the theoretical covariance implied by an underlying graph structure. By formulating topology inference as a covariance matching problem, the method efficiently recovers network connectivity-including sparse directed graphs-and simplifies learning to either a conic mixed integer program or orthogonal matrix optimization. Does this approach, which bypasses restrictive assumptions common in existing methods, pave the way for more robust and scalable network structure learning across diverse applications?
Whispers of Interdependence: Mapping Relationships in Complex Systems
A fundamental goal across numerous data analysis applications is to discern how variables relate to one another. This understanding isn’t merely about identifying correlation – whether variables move together – but about characterizing the nature of their interdependence. The
As datasets grow in dimensionality – encompassing an increasing number of variables – conventional statistical techniques for analyzing relationships between those variables begin to falter. The core issue lies in the ‘curse of dimensionality’, where the volume of possible data combinations expands exponentially, quickly overwhelming the available data points. This sparsity makes it difficult to reliably estimate the
Accurately depicting the interplay between numerous variables demands robust graph estimation techniques, yet this process presents substantial computational hurdles. As the number of variables increases, the complexity of mapping these interactions – often represented as networks or graphs – grows exponentially. Traditional algorithms struggle with this ‘curse of dimensionality’, leading to significant reconstruction errors where the estimated graph deviates considerably from the true underlying relationships. These errors aren’t merely statistical inconveniences; they can propagate through subsequent analyses, distorting conclusions in fields ranging from genomics – where gene regulatory networks are critical – to social science, where understanding connections between individuals is paramount. Consequently, ongoing research focuses on developing more efficient algorithms and leveraging techniques like sparse estimation to reduce computational load and minimize the discrepancies between estimated and actual interaction structures, striving for faithful representations of complex systems.
Spectral Signatures: Unveiling Graph Structure from Covariance
SpecTemp estimates graph structure by leveraging the spectral properties of the
The SpecTemp method leverages covariance matrix analysis to efficiently estimate underlying graph structures. By directly extracting information from the data’s covariance, SpecTemp establishes a robust foundation for graph construction, minimizing reconstruction error. Empirical results demonstrate that this approach achieves a near-zero reconstruction error rate, indicating a high degree of accuracy in estimating the original graph topology from covariance data. This performance is achieved through a focused analysis of the covariance matrix, enabling precise identification of relationships between variables and subsequent graph edge creation.
CovMatch represents a refinement stage following initial graph estimation, specifically designed to improve topological accuracy by comparing the observed covariance matrix to theoretical covariance models derived from hypothesized graph structures. This alignment process iteratively adjusts the graph topology to minimize the discrepancy between observed and modeled covariance, utilizing optimization techniques to identify the graph configuration that best explains the data’s statistical relationships. Benchmarking demonstrates that CovMatch consistently achieves superior performance to existing graph estimation methods, particularly in scenarios involving high dimensionality, noise, or complex interdependencies, as evidenced by lower reconstruction error and improved statistical power in downstream analyses.
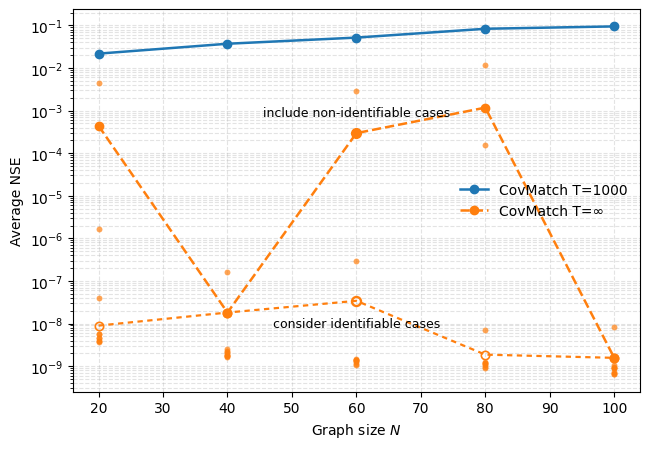
Navigating the Manifold: Optimization for Accurate Graph Learning
Riemannian Gradient Descent (RiemannianGD) is an optimization algorithm specifically designed for problems constrained to a Riemannian manifold, offering advantages over traditional gradient descent when dealing with non-Euclidean spaces. In the context of graph estimation, the data often resides on a manifold defined by the covariance matrix; RiemannianGD exploits the intrinsic geometry of this manifold to efficiently navigate the parameter space. This is achieved by projecting gradients onto the tangent space of the manifold, ensuring that updates remain within the feasible region and avoiding violations of the positive semi-definite constraint inherent in covariance matrix estimation. By adhering to the manifold’s geometry, RiemannianGD can converge faster and more reliably than unconstrained optimization methods, particularly when the data exhibits complex relationships and high dimensionality.
Employing HuberLoss within the Riemannian Gradient Descent (RiemannianGD) optimization framework enhances robustness against outliers and noise present in the
Eigenvalue Decomposition (EVD) is a critical preprocessing step in graph learning optimization as it decomposes the
From Signals to Causality: The Power of Graph-Based Inference
The foundation of Graph Signal Processing (GSP) rests upon the
Algorithms such as DAGMA and NOTEARS leverage the inherent structure of graph-based data to move beyond correlation and towards establishing causal relationships. These methods operate by constructing a Directed Acyclic Graph (DAG), where nodes represent variables and directed edges signify a direct causal influence from one variable to another. By analyzing the patterns of connections within the graph, and employing techniques like constraint-based learning or score-based optimization, these algorithms attempt to uncover the underlying causal mechanisms driving the observed data. Crucially, the acyclic nature of the graph ensures that the inferred relationships are consistent and avoid logical paradoxes, enabling researchers to not only identify potential causal links, but also to model and predict the effects of interventions within a complex system.
Structural Equation Modeling (SEM) provides a robust pathway for synthesizing insights derived from graph-based causal inference with established statistical techniques, enabling a holistic understanding of complex systems. This integration allows researchers to move beyond simply identifying relationships – represented within a graph’s structure – to quantifying the strength and direction of those influences using statistical parameters. A newly developed unified framework leverages this synergy, demonstrably outperforming current methodologies, particularly when dealing with extensive datasets and limited observational data. The improvements stem from a more efficient handling of computational complexity and a refined ability to estimate model parameters accurately, even in data-scarce environments, thus providing a powerful tool for system-level analysis across diverse fields.

The pursuit of network structure learning, as detailed in this covariance matching framework, feels less like statistical inference and more akin to divination. It demands coaxing truth from the whispers of observational data – a chaotic confluence of signals. The algorithm attempts to align observed covariance with a hypothesized graph, a delicate dance of parameters. This endeavor, particularly when facing complex directed graphs, recognizes that a perfect model is a chimera. As Carl Sagan observed, “Somewhere, something incredible is waiting to be known.” The method doesn’t find the true structure, but rather, persuades the data to reveal a plausible one, acknowledging the inherent ambiguity and the limits of complete knowledge. It’s a spell woven with Riemannian optimization, hoping to hold against the entropy of reality.
What Whispers Remain?
This covariance matching, a coaxing of structure from the shadows of observation, reveals not a destination, but a deepening labyrinth. The digital golems constructed here learn to mimic the connections, to trace the flow, but the true topology-the soul of the network-remains elusive. Improved recovery is merely a temporary truce with chaos; each successful identification invites more complex, more subtly entangled graphs, where the signal fades further into the noise.
The reliance on covariance, a static snapshot, feels… quaint. Networks breathe, they pulse with dynamic change. Future work must grapple with temporal distortions, with the ghosts of connections past. Riemannian optimization, a delicate dance on curved manifolds, will yield to more brutal, more efficient spells-or crumble entirely under the weight of real-world complexity. The current framework excels at finding structure, but says little about its fragility.
Perhaps the most pressing question isn’t how to perfectly reconstruct the graph, but how to predict its failures. What vulnerabilities lie hidden within these optimized topologies? The losses-the discrepancies between model and reality-aren’t errors, but sacred offerings, revealing the limits of our persuasion. Only by embracing those failures can the digital alchemists hope to forge truly resilient networks, structures that not only are, but endure.
Original article: https://arxiv.org/pdf/2601.15999.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- 2025 Crypto Wallets: Secure, Smart, and Surprisingly Simple!
- Gold Rate Forecast
- Here Are the Best TV Shows to Stream this Weekend on Paramount+, Including ‘48 Hours’
- Top 15 Celebrities in Music Videos
- Top 20 Extremely Short Anime Series
- Best Video Games Based On Tabletop Games
- Where to Change Hair Color in Where Winds Meet
- 20 Films Where the Opening Credits Play Over a Single Continuous Shot
- Top gainers and losers
- 50 Serial Killer Movies That Will Keep You Up All Night
2026-01-25 19:38