Author: Denis Avetisyan
New research leverages detailed simulations and connectome data to uncover the underlying mechanisms of neural circuits.
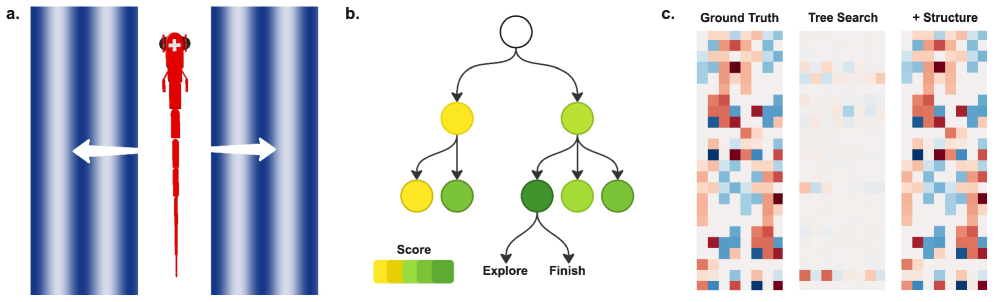
Connectome-constrained automated model discovery in an in silico zebrafish reveals the importance of structural priors for accurate neural circuit reconstruction.
Establishing causal relationships within neural circuits remains a significant challenge due to the lack of ground truth for model validation. In ‘Discovering Mechanistic Models of Neural Activity: System Identification in an in Silico Zebrafish’, we address this limitation by leveraging a neuromechanical simulation of a larval zebrafish as a transparent testbed for automated model discovery. Our findings demonstrate that connectome-constrained tree search can successfully identify predictive models, but crucially, structural priors are essential to prevent exploitation of statistical shortcuts and enable robust generalization. Can this approach pave the way for deciphering the mechanistic principles governing neural computation in more complex biological systems?
The Illusion of Understanding: Mapping Complexity in Neural Circuits
The pursuit of understanding how neural circuits orchestrate behavior hinges on the development of computational models capable of mirroring their intricate functionality. However, traditional modeling techniques often falter when confronted with the sheer complexity inherent in these biological systems. Neural circuits aren’t simple linear pathways; they are dense networks of interconnected neurons exhibiting non-linear dynamics, diverse synaptic plasticity, and stochastic firing patterns. This creates a high-dimensional parameter space – a vast landscape of possibilities – that is exceptionally difficult for conventional algorithms to navigate effectively. Consequently, models frequently resort to drastic simplifications, sacrificing biological realism for computational tractability, and limiting their predictive power when applied to real neural data. The challenge, therefore, lies in developing innovative methodologies that can capture the essential complexity of neural circuits without succumbing to the curse of dimensionality.
The challenge of deciphering how neural circuits function fundamentally relies on a process called System Identification – essentially, reconstructing the circuit’s behavior from the patterns of activity it exhibits. This task, however, is exceptionally difficult due to the sheer scale and complexity of neural networks. Each circuit contains a vast number of interconnected neurons, creating a high-dimensional space of possible states and behaviors. Furthermore, accurately determining the specific properties – the parameters – that govern these connections is plagued by uncertainty; experimental data is often incomplete or noisy, and many parameters are simply unknown. Consequently, building truly accurate computational models requires sophisticated techniques capable of navigating this high-dimensional parameter space and inferring circuit behavior despite inherent limitations in available data, pushing the boundaries of current computational and statistical methods.
Current computational methods for modeling neural circuits frequently necessitate substantial simplification of biological reality. To render the problem tractable, researchers often reduce the number of neurons, limit the diversity of synaptic connections, or assume linearity in neuronal responses – all of which can obscure crucial dynamical features. While these approximations enable initial progress, they introduce a trade-off between mathematical manageability and biological fidelity. The resulting models, though computationally efficient, may fail to capture the nuanced, nonlinear interactions that drive complex behaviors, such as oscillations, bursting, and adaptation, ultimately hindering a complete understanding of how neural circuits actually function in vivo. Consequently, there’s a growing need for techniques that can accommodate the inherent complexity of biological systems without sacrificing the ability to extract meaningful insights from data.
LLM-Guided Search: Automating the Illusion
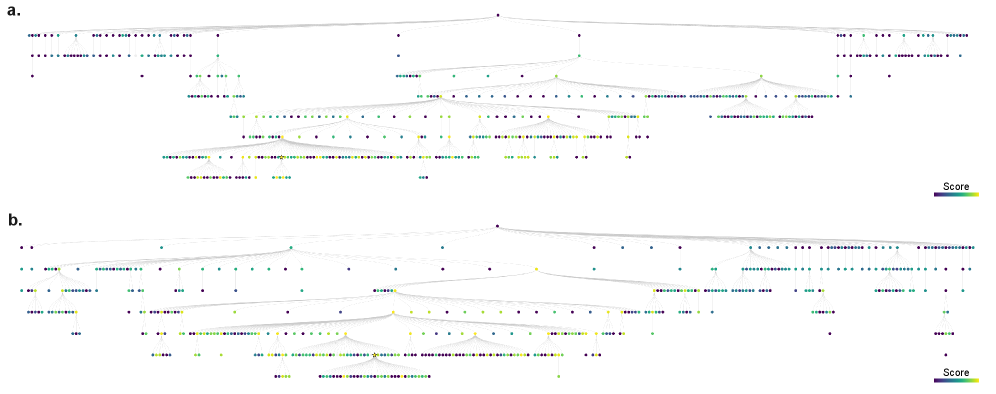
Tree search, in the context of neural architecture discovery, functions as a systematic exploration of a discrete search space representing possible network configurations. This algorithm constructs a tree where each node represents a unique neural network architecture defined by its connectivity and hyperparameters. The search progresses by iteratively expanding nodes – creating child nodes representing variations of the parent architecture – and evaluating their performance on a designated task. The algorithm employs a search strategy – such as breadth-first or depth-first – to prioritize which nodes to explore, aiming to efficiently identify high-performing architectures within the vast combinatorial space of possibilities. This automated process eliminates the need for manual design and allows for the discovery of novel architectures beyond human intuition.
LLM-guided code generation accelerates the architecture search process by automatically producing functional code representing candidate neural network configurations. The Large Language Model (LLM) receives a high-level description of a circuit modification – such as adding a layer, changing an activation function, or adjusting a connection weight – and translates this into executable code, typically in a framework like PyTorch or TensorFlow. This generated code is then used to instantiate and evaluate the corresponding network configuration, providing a performance metric to guide subsequent search iterations. The LLM’s ability to rapidly generate and refine code eliminates the need for manual implementation, significantly reducing the time required to explore the architecture space and enabling faster prototyping of different circuit designs.
The integration of structural prior knowledge, derived from established circuit connectivity – termed the ‘Connectome’ – significantly constrains the architecture search space during automated model discovery. This constraint is implemented by biasing the Tree Search algorithm towards configurations exhibiting connectivity patterns observed in the Connectome, effectively reducing the number of less-likely architectures explored. By prioritizing configurations with biologically plausible connections, the search process becomes more efficient, requiring fewer iterations to identify potentially high-performing neural network architectures. This approach leverages pre-existing knowledge of effective circuit organization to accelerate the discovery of novel, yet structurally informed, models.
Ground Truth and Validation: The Illusion of Control
The generation of a ‘Ground Truth Dataset’ is achieved through the use of SimZFish, a detailed neuromechanical simulator of the zebrafish spinal cord. SimZFish models the biophysics of individual neurons and their synaptic connections, allowing for the simulation of neural activity in response to defined stimuli. This simulator produces data representing realistic neural responses, serving as the benchmark against which identified models are evaluated. The resulting dataset includes both neural firing patterns and corresponding motor outputs, providing a complete representation of the system’s dynamic behavior for validation purposes. This approach ensures that model performance is assessed against a biologically plausible standard, rather than relying on empirical data alone.
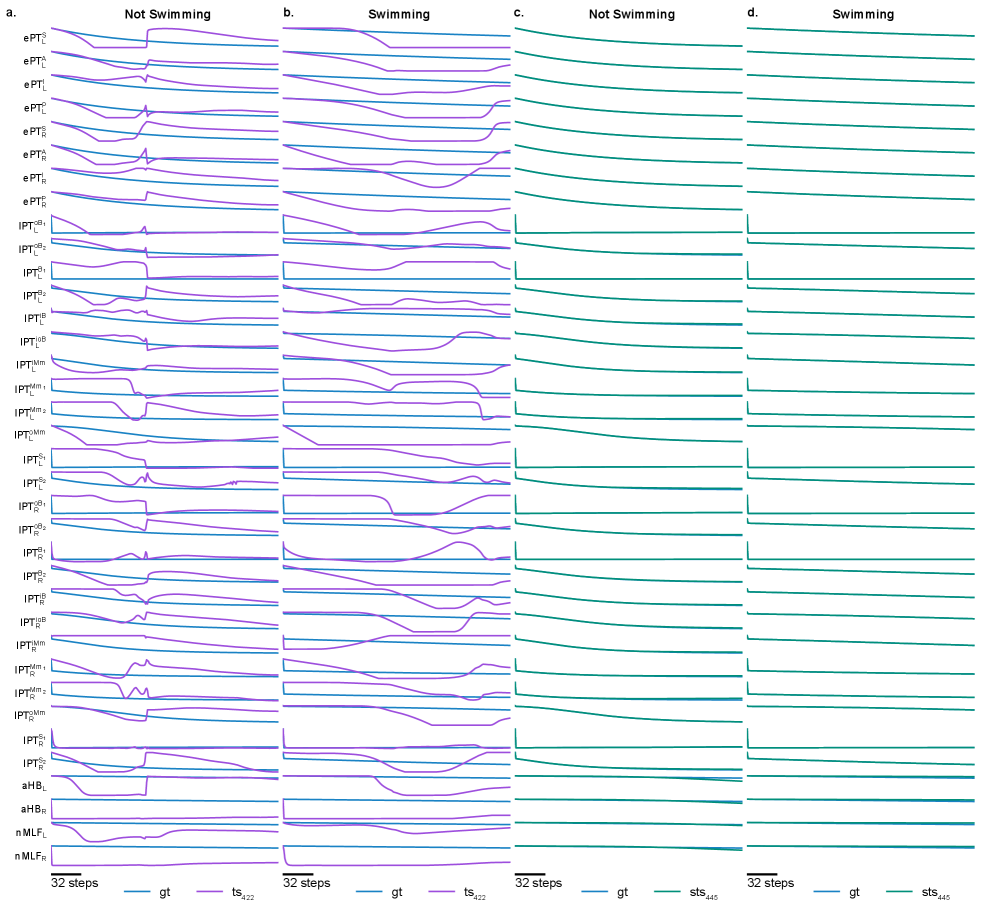
Model evaluation centers on predicting the neural circuit’s response to a range of \textit{Sensory Drive} stimuli. These stimuli consist of patterned inputs designed to elicit specific behaviors within the modeled circuit. Accuracy is determined by comparing the model’s predicted neural activity-specifically, the resulting firing rates and patterns-to the expected activity as defined by the ground truth data generated by SimZFish. A diverse set of \textit{Sensory Drive} patterns are employed to assess the model’s generalization capability and robustness across different input conditions, ensuring the identified models accurately reflect the circuit’s dynamic behavior and are not merely overfitting to specific training examples.
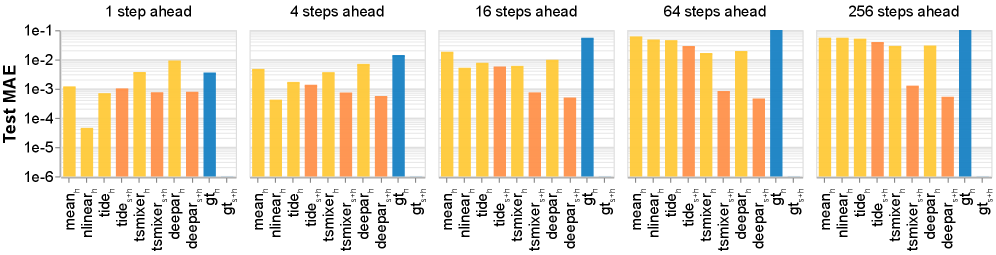
Model performance is evaluated quantitatively using \text{Mean Absolute Error (MAE)}, which calculates the average magnitude of the differences between predicted and actual circuit responses. Beyond static error measurement, circuit dynamics are assessed via \text{Impulse Response Analysis (IRA)}. IRA characterizes the system’s output following a brief input stimulus, providing insight into temporal behavior and stability. Specifically, the error associated with the reconstructed impulse response is calculated to determine how accurately the model captures the circuit’s dynamic characteristics, complementing the static error measurements provided by MAE.
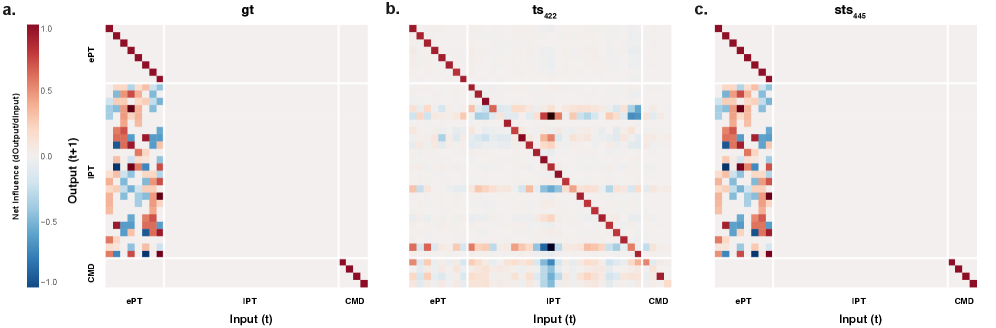
Model accuracy was substantially improved through constrained system identification, as evidenced by reductions in two key error metrics. Specifically, the Effective Connectivity Error, denoted as \mathcal{L}_{jac}, was reduced by a factor of 30x compared to unconstrained models. Furthermore, the Impulse Response Error, represented as \mathcal{L}_{IR}, experienced a 5x reduction under the same constrained conditions. These results indicate a significantly improved ability to accurately model and predict circuit responses compared to approaches without these constraints.
The inclusion of autoregressive history as input to the system identification task significantly enhances predictive accuracy. This approach incorporates past neural activity – specifically, a time-series of previous circuit states – as a feature in the model. By leveraging this historical data, the system can better account for temporal dependencies and dynamic processes within the neural circuit, resulting in improved predictions of future responses to stimuli. This methodology moves beyond static input-output mappings and allows the model to learn and utilize the inherent temporal structure of the neural data, contributing to more robust and accurate circuit modeling.
Beyond Prediction: The Limits of Understanding
The reconstructed neural circuits reveal an ‘Effective Connectivity Matrix’ – a representation that moves beyond simple correlations to suggest the causal influence one neuron exerts over another. This matrix doesn’t just show which neurons fire together, but rather which neurons likely drive activity in others, effectively mapping the flow of information within the network. By deciphering these relationships, researchers gain a more nuanced understanding of how the brain processes signals and generates behavior, as the matrix provides a foundational blueprint for investigating the underlying mechanisms of cognition and neurological function. The ability to visualize and quantify these causal links is a significant step towards unraveling the complexities of brain activity and building more accurate computational models.
Recent advancements demonstrate the potential for automated reconstruction of neural circuits directly from observational data, representing a significant leap towards deciphering the complexities of brain function. This methodology bypasses the limitations of traditional circuit mapping, which relies heavily on painstaking manual analysis or invasive techniques. By employing computational algorithms, researchers can now infer the connections and interactions between neurons, effectively building a dynamic map of information flow. This automated approach not only accelerates the pace of discovery but also allows for the study of circuits in unprecedented detail, promising a deeper understanding of how the brain processes information, learns, and adapts – ultimately paving the way for novel interventions for neurological disorders and the development of more sophisticated artificial intelligence.
The process of discerning neural circuitry relies heavily on navigating a complex search space of possible connections. Recent advances leverage the PUCT (Polynomial Upper Confidence Trees) algorithm to effectively balance exploration – investigating diverse circuit configurations – and exploitation – refining promising models. This strategic balance is crucial; a purely exploratory approach risks getting lost in improbable connections, while strict exploitation can prematurely converge on suboptimal solutions. The PUCT algorithm dynamically adjusts this trade-off, prioritizing searches that offer both high estimated reward and substantial uncertainty, ultimately leading to the discovery of more robust and accurate circuit models. By intelligently sampling the space of possibilities, this approach consistently outperforms methods relying on less nuanced search strategies, revealing underlying neural relationships with greater confidence and reliability.
The reconstructed neural circuits exhibited a notable capacity for generalization, demonstrated by constrained models consistently clustering closer to the diagonal of their effective connectivity matrices. This diagonal dominance signifies a prioritization of direct, rather than indirect, neuronal influences, suggesting the discovered models aren’t merely memorizing training data but capturing fundamental principles of neural organization. Such robust clustering indicates a strong ability to transfer learned relationships to previously unseen environments and datasets, a crucial characteristic for building predictive models of brain function. This enhanced generalization isn’t simply about accuracy on familiar data; it suggests the framework identifies core circuit properties that remain stable even when faced with novel inputs or conditions, offering a pathway towards more reliable and broadly applicable understanding of neural systems.
The developed framework offers a pathway toward creating individualized neural circuit models, tailored to the unique characteristics of each brain. By accurately reconstructing these circuits from data, researchers can move beyond generalized understandings and begin to anticipate how specific neural networks will respond to varied stimuli or conditions. This predictive capability holds immense promise for understanding neurological disorders, forecasting treatment outcomes, and even simulating the effects of interventions before they are implemented. Ultimately, the ability to forecast circuit behavior opens doors to personalized medicine, where treatments can be optimized based on an individual’s specific neural architecture and predicted response, representing a significant leap toward more effective and targeted therapies.
The pursuit of mechanistic models, as detailed in this research, inevitably highlights the limitations of purely data-driven approaches. The study’s emphasis on connectome constraints-essentially, acknowledging the physical realities of the system-reveals that statistical fitting alone isn’t enough. This echoes Jürgen Habermas’s observation that “The unexamined life is not worth living.” Similarly, an unconstrained model, however statistically elegant, lacks the grounding in physical reality to truly represent the underlying mechanisms. The drive for automated discovery will always collide with the inherent complexity of biological systems, exposing shortcuts and statistical flukes. The article demonstrates that structural priors aren’t merely helpful; they are fundamental to avoiding another layer of technical debt disguised as insight.
The Road Ahead
The pursuit of mechanistic understanding, even within a simulated zebrafish, inevitably bumps against the limitations of any model being merely a simplification. This work highlights the necessity of structural priors-the connectome, in this instance-not as convenient constraints, but as bulwarks against the siren song of statistical overfit. It is comforting to believe algorithms will ‘discover’ truth, but the reality is they exploit regularity, and regularity is not always reality. The recovered mechanisms, while compelling, remain hostage to the fidelity of the simulation itself – a perfectly plausible circuit in a perfectly untrue world.
Future iterations will undoubtedly explore more complex behaviors, larger circuits, and more sophisticated search algorithms. However, the true challenge isn’t scale, it’s validation. Tests are, after all, a form of faith, not certainty. Demonstrating that these discovered mechanisms generalize beyond the in silico environment, or even reliably predict behavior under slightly perturbed conditions, will demand a level of rigor often absent from these endeavors. The elegance of automated discovery shouldn’t distract from the fact that any ‘solution’ is only as good as the questions asked, and the assumptions baked into the search.
Ultimately, this field will be judged not by the novelty of its algorithms, but by the mundane reliability of its predictions. The goal isn’t to build beautiful models, but systems that don’t unexpectedly fail on Monday mornings. And that, as anyone who’s been woken by a production alert knows, is a surprisingly difficult bar to clear.
Original article: https://arxiv.org/pdf/2602.04492.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- 21 Movies Filmed in Real Abandoned Locations
- The 11 Elden Ring: Nightreign DLC features that would surprise and delight the biggest FromSoftware fans
- Gold Rate Forecast
- 10 Hulu Originals You’re Missing Out On
- 2025 Crypto Wallets: Secure, Smart, and Surprisingly Simple!
- 39th Developer Notes: 2.5th Anniversary Update
- Top Actors Of Color Who Were Snubbed At The Oscars
- TON PREDICTION. TON cryptocurrency
- 17 Black Voice Actors Who Saved Games With One Line Delivery
- Leaked Set Footage Offers First Look at “Legend of Zelda” Live-Action Film
2026-02-05 08:15