Author: Denis Avetisyan
New research explores how connecting large language models with focused knowledge bases improves their ability to answer complex questions about diseases like Alzheimer’s and Type 2 Diabetes.
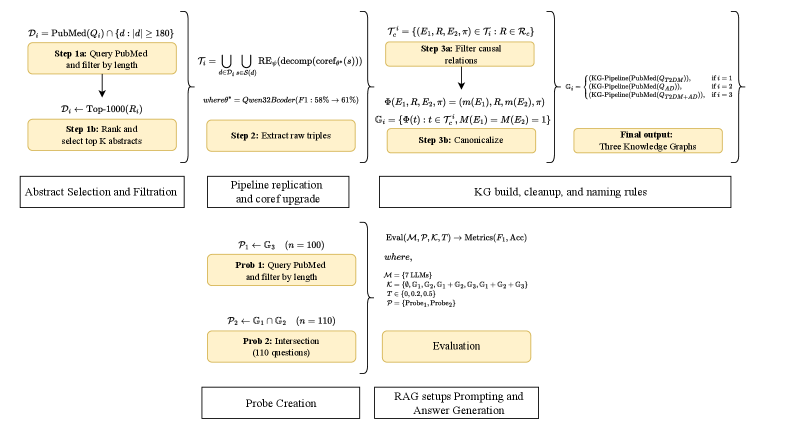
This review examines the impact of domain-specific knowledge graphs on the performance of retrieval-augmented generation systems in biomedical applications.
While large language models excel at generating fluent text, ensuring trustworthy reasoning in specialized domains remains a challenge. This is addressed in ‘Domain-Specific Knowledge Graphs in RAG-Enhanced Healthcare LLMs’, which investigates the impact of curated biomedical knowledge graphs on retrieval-augmented generation (RAG) for complex queries related to Alzheimer’s disease and Type 2 Diabetes. The research demonstrates that precise alignment between the scope of a knowledge graph and the target query is crucial-well-defined graphs yield consistent gains, whereas indiscriminate unions can introduce noise-and that the benefits of RAG are more pronounced for smaller language models leveraging strong parametric priors. How can we best design and integrate domain-specific knowledge to maximize the reliability and accuracy of LLM-powered healthcare applications?
The Convergence of Neurological and Metabolic Dysfunction
The growing recognition of shared pathological mechanisms between Alzheimer’s Disease and Type 2 Diabetes Mellitus challenges the traditional view of these conditions as distinct entities. Once considered separate neurological and metabolic disorders, research now demonstrates converging pathways, particularly concerning insulin resistance and impaired glucose metabolism within the brain. This phenomenon, sometimes referred to as ‘Type 3 Diabetes’, suggests that disruptions in peripheral glucose homeostasis can directly impact neuronal function and contribute to the amyloid plaque and tau tangle formation characteristic of Alzheimer’s. Consequently, the brain’s ability to utilize glucose, essential for energy and synaptic plasticity, becomes compromised, accelerating cognitive decline. This convergence isn’t merely correlational; studies reveal similar inflammatory responses and cellular stress markers in both diseases, pointing to a fundamental, systemic link that may redefine diagnostic and therapeutic strategies for both conditions.
The convergence of Alzheimer’s Disease and Type 2 Diabetes isn’t merely coincidental; both conditions demonstrably involve persistent, low-grade inflammation and an imbalance between the production of free radicals and the body’s ability to neutralize them – a state known as oxidative stress. This shared pathophysiology suggests a systemic connection extending beyond localized effects. Chronic inflammation disrupts cellular processes crucial for both glucose metabolism and neuronal function, while oxidative stress damages proteins, lipids, and DNA, contributing to insulin resistance and the formation of amyloid plaques characteristic of Alzheimer’s. Research indicates these processes aren’t isolated to the brain or pancreatic cells, but rather represent a broader systemic dysfunction, potentially initiated by factors like aging, diet, and lifestyle, ultimately increasing vulnerability to both debilitating diseases.
The convergence of Alzheimer’s disease and type 2 diabetes extends beyond shared inflammatory pathways to encompass significant genetic overlaps, most notably the Apolipoprotein E4 (APOE4) gene. This gene, instrumental in cholesterol metabolism and neuronal lipid transport, represents a substantial risk factor for both conditions; individuals carrying the APOE4 allele exhibit a markedly increased susceptibility to developing either Alzheimer’s or type 2 diabetes compared to those with other APOE variants. Research suggests APOE4 impairs insulin signaling in the brain and periphery, contributing to glucose dysregulation, while simultaneously hindering amyloid-beta clearance, a hallmark of Alzheimer’s pathology. The consistent association between APOE4 and both diseases provides compelling evidence for shared underlying vulnerabilities and suggests that interventions targeting this genetic predisposition could potentially offer preventative or therapeutic benefits for both Alzheimer’s and type 2 diabetes.
Constructing a Cognitive Map: A Knowledge Graph Approach
Probe 1 is a knowledge-driven system specifically designed to investigate the complex interplay between Alzheimer’s Disease and Type 2 Diabetes, moving beyond the identification of instances where both conditions are simply mentioned together. The system aims to identify and characterize relationships – such as shared genetic factors, common pathological mechanisms, or the influence of diabetes on Alzheimer’s progression – that explain why these two conditions frequently co-occur. This is achieved through knowledge graph analysis, allowing for the inference of connections that are not explicitly stated in the source literature, and providing a more nuanced understanding of their relationship than traditional co-occurrence studies.
Knowledge Graphs represent biomedical information by modeling entities – such as genes, diseases, and drugs – as nodes and the relationships between them as edges. This structure moves beyond simple keyword searches or co-occurrence analysis, allowing the system to infer connections and patterns not explicitly stated in source texts. Specifically, relationships are defined by semantic types – for example, ‘disease_of’, ‘treats’, or ‘interacts_with’ – providing context for each connection. This interconnected representation enables complex reasoning, such as identifying potential drug repurposing candidates or predicting disease progression pathways, by traversing the graph and applying logical rules or statistical analysis to the network of relationships.
The CoDe-KG Pipeline employs natural language processing techniques, including entity recognition, relation extraction, and coreference resolution, to automatically construct Knowledge Graphs from unstructured biomedical text. This automated approach significantly reduces the time and resources traditionally required for manual curation of biomedical knowledge. The pipeline integrates publicly available databases, such as PubMed and clinical trial repositories, as data sources and utilizes a standardized ontology to ensure consistency and interoperability within the resulting graph. By automating graph construction, the CoDe-KG Pipeline enables rapid iteration and scaling of knowledge discovery efforts, facilitating the identification of previously unknown relationships between diseases, genes, and treatments.
Inferring Connections: Navigating the Network of Disease
Probe 1 employs both Single-Hop Relation and Multi-Hop Relation inference to identify connections between seemingly unrelated diseases. Single-Hop Relation inference directly links two concepts based on a known relationship, while Multi-Hop Relation inference establishes a connection by traversing intermediate concepts and relationships within a knowledge network. This allows the system to uncover associations that are not immediately apparent, revealing potential pathways between diseases that may not be readily identified through conventional research methods. The system’s ability to utilize both methods expands its capacity for identifying complex relationships and uncovering previously unappreciated links.
The system can identify relationships between seemingly unrelated conditions, such as Insulin Resistance, a key feature of Type 2 Diabetes, and Tau Tangles, a pathological hallmark of Alzheimer’s Disease. This connection is not direct; rather, it is established through the identification of intermediate concepts and relationships. These intermediate concepts act as a bridge, revealing a pathway linking the physiological processes underlying both conditions. For instance, the system might identify shared molecular pathways or risk factors that connect Insulin Resistance to the development or progression of Tau Tangles, thereby demonstrating a non-obvious association.
The system’s network traversal capability extends analytical reasoning beyond immediately obvious relationships between concepts. Rather than being limited to direct associations, the system identifies connections through intermediate nodes and pathways within a knowledge graph. This allows for the discovery of non-obvious relationships, such as those linking seemingly unrelated diseases, by synthesizing information from multiple sources and inferring connections based on complex relational patterns. The ability to navigate these networks facilitates a deeper understanding of underlying mechanisms and potential shared factors that would not be apparent through simple keyword matching or direct association analysis.
Grounding Language Models in Verified Knowledge: A Paradigm Shift
The integration of Probe 1 with Retrieval-Augmented Generation (RAG) represents a significant step towards more reliable and informed responses from Large Language Models. This approach allows models to move beyond simply generating text based on learned patterns and instead access a structured Knowledge Graph to retrieve relevant, verified information. By supplementing the model’s inherent knowledge with external data, RAG effectively anchors responses in factual accuracy, mitigating the risk of hallucinations or the propagation of misinformation. The process involves identifying key concepts within a user’s query and then searching the Knowledge Graph for corresponding data, which is then incorporated into the model’s response generation. This dynamic access to curated knowledge not only improves the trustworthiness of the output but also enables the model to answer questions requiring specialized or up-to-date information that may not have been part of its original training data.
Large Language Models, while impressively versatile, are fundamentally prone to generating outputs that, while seemingly coherent, lack factual grounding – a phenomenon often referred to as ‘hallucination’. This arises from their predictive text-generation architecture; they excel at statistically likely sequences but don’t inherently possess a mechanism to verify truthfulness. Consequently, LLMs can confidently present inaccurate or entirely fabricated information. Integrating structured knowledge sources, as demonstrated by this work, directly addresses this limitation by providing a reliable external reference point. This grounding process constrains the model’s output to verified facts, dramatically reducing the incidence of hallucinations and bolstering the overall reliability of its responses – a crucial step towards deploying these powerful tools in domains demanding precision and trustworthiness.
The integration of a knowledge graph with large language models demonstrably elevates response accuracy, reaching 99% with leading models when evaluated on Probe 1. This substantial improvement arises from grounding the models in verified information, effectively mitigating the risk of generating inaccurate or fabricated content. Further analysis reveals significant gains in F1 scores for Mixtral-8x7B on Probe 2, achieved across both Alzheimer’s Disease-focused knowledge graphs (𝔾2) and combined Alzheimer’s Disease and Type 2 Diabetes Mellitus graphs (𝔾1+𝔾2). These results underscore the effectiveness of this approach in complex domains, providing a robust mechanism for ensuring the reliability and trustworthiness of generated text.
Evaluations revealed a substantial performance disparity between models when presented with complex, intersection-style questions in Probe 2; Gemini experienced a 36.3% accuracy decrease, while ChatGPT’s accuracy dropped by 28%. This significant decline underscores a critical limitation of Large Language Models when reasoning across multiple knowledge domains without external support. The results emphasize that simply scaling model parameters isn’t sufficient to overcome challenges requiring nuanced connections between disparate facts; instead, a meticulously designed knowledge graph serves as a foundational element for accurate and reliable responses. The observed accuracy drop demonstrates that a well-structured knowledge graph isn’t merely a supplemental tool, but a necessity for LLMs attempting to navigate and synthesize information from complex, interconnected datasets.
The pursuit of robust reasoning within large language models, as demonstrated by this research into knowledge graphs and RAG systems, echoes a fundamental principle of computational elegance. It’s not merely about achieving a functional output, but establishing provable correctness. As Marvin Minsky stated, “The more we understand about how the brain works, the more we realize how much we don’t.” This sentiment underscores the necessity of meticulously constructed knowledge graphs-specifically tailored to complex domains like Alzheimer’s Disease and Type 2 Diabetes-to augment LLMs. The study’s focus on domain-specific knowledge isn’t simply about improving answer accuracy; it’s about building a foundation of logical completeness, ensuring the system’s reasoning isn’t a matter of statistical probability but of demonstrable truth.
The Road Ahead
The demonstrated influence of curated knowledge graphs on retrieval-augmented generation systems, while promising, merely highlights the persistent chasm between statistical correlation and genuine understanding. The current reliance on biomedical information extraction techniques to populate these graphs remains a fundamentally brittle endeavor. A system’s capacity to answer complex queries is only as robust as its ability to consistently and accurately represent underlying biological truths – a feat far beyond the reach of pattern recognition. The temptation to simply scale graph size, as a proxy for knowledge depth, is a particularly insidious distraction.
Future work must address the limitations of relying on text-derived relationships. A shift toward knowledge representation grounded in formal logic, perhaps incorporating causal Bayesian networks, offers a potential, if arduous, path toward building systems capable of deductive reasoning, rather than sophisticated memorization. The elegance of an algorithm, after all, resides not in its ability to parrot information, but in the consistency of its boundaries and predictability – a principle too often obscured by the allure of large datasets.
The ultimate benchmark will not be performance on standardized question-answering tasks, but rather the capacity to generate novel, testable hypotheses. Until these systems can reliably move beyond recombination of existing knowledge, they remain, at best, elaborate search engines – impressive, certainly, but fundamentally incapable of true scientific discovery.
Original article: https://arxiv.org/pdf/2601.15429.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- 39th Developer Notes: 2.5th Anniversary Update
- The 10 Most Beautiful Women in the World for 2026, According to the Golden Ratio
- TON PREDICTION. TON cryptocurrency
- Gold Rate Forecast
- Bitcoin’s Bizarre Ballet: Hyper’s $20M Gamble & Why Your Grandma Will Buy BTC (Spoiler: She Won’t)
- Nikki Glaser Explains Why She Cut ICE, Trump, and Brad Pitt Jokes From the Golden Globes
- Ephemeral Engines: A Triptych of Tech
- Dividends: A Most Elegant Pursuit
- Venezuela’s Oil: A Cartography of Risk
- AI Stocks: A Slightly Less Terrifying Investment
2026-01-25 17:59