Author: Denis Avetisyan
Researchers have developed a new deep learning method to generate realistic dark-field radiographs from standard X-ray images, unlocking potentially valuable diagnostic information.
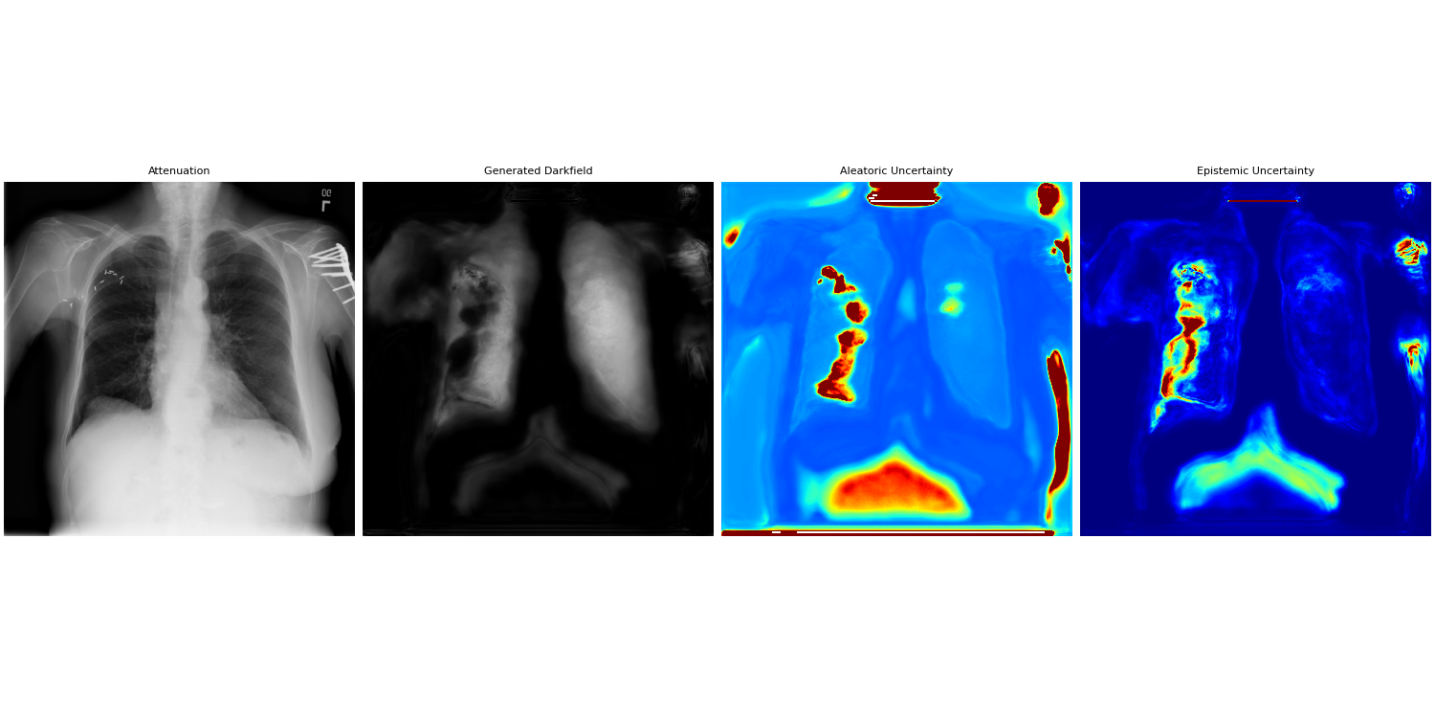
This work introduces a GAN-based framework incorporating both aleatoric and epistemic uncertainty quantification to improve the reliability and interpretability of synthetic dark-field imaging.
While X-ray dark-field radiography offers unique microstructural insights complementary to conventional imaging, its limited data availability hinders the development of robust deep learning applications. This work, ‘Uncertainty-guided Generation of Dark-field Radiographs’, introduces a novel framework leveraging generative adversarial networks to synthesize realistic dark-field images directly from standard attenuation radiographs. By explicitly modeling both aleatoric and epistemic uncertainty, the approach enhances the reliability and interpretability of generated images, demonstrating improved fidelity and generalization. Could this uncertainty-guided synthesis pave the way for wider clinical adoption and advanced diagnostic capabilities in dark-field imaging?
Beyond Simple Density: Revealing Hidden Tissue Detail
Conventional radiography relies on measuring how much X-ray radiation is absorbed as it passes through the body, creating images based on density differences. While adept at visualizing bones and identifying large-scale abnormalities, this attenuation-based approach often fails to capture the subtle changes occurring within tissues at a microstructural level. Crucially, early-stage diseases, or even normal physiological shifts, may manifest as alterations in tissue architecture – changes in the arrangement of cells, fibers, or fluid distribution – that produce minimal differences in overall density. Consequently, vital information regarding tissue health and function can remain hidden, limiting diagnostic accuracy and potentially delaying critical interventions. The technique essentially provides a ‘bulk’ measurement, overlooking the intricate details that define a tissue’s true condition and responsiveness to disease or therapy.
Conventional radiography, despite its widespread use, faces inherent limitations when it comes to identifying subtle pathological changes within tissues. The technique primarily relies on measuring X-ray attenuation – how much radiation is blocked by a material – which excels at visualizing dense structures like bone or tumors. However, many diseases manifest as more delicate alterations in tissue microstructure before becoming visibly dense, escaping detection by standard methods. This poses a significant diagnostic challenge, particularly in early-stage disease where intervention is most effective. Consequently, relying solely on attenuation-based imaging can lead to delayed diagnoses or a failure to fully characterize the extent of disease, underscoring the need for complementary imaging modalities capable of revealing these nuanced tissue characteristics.
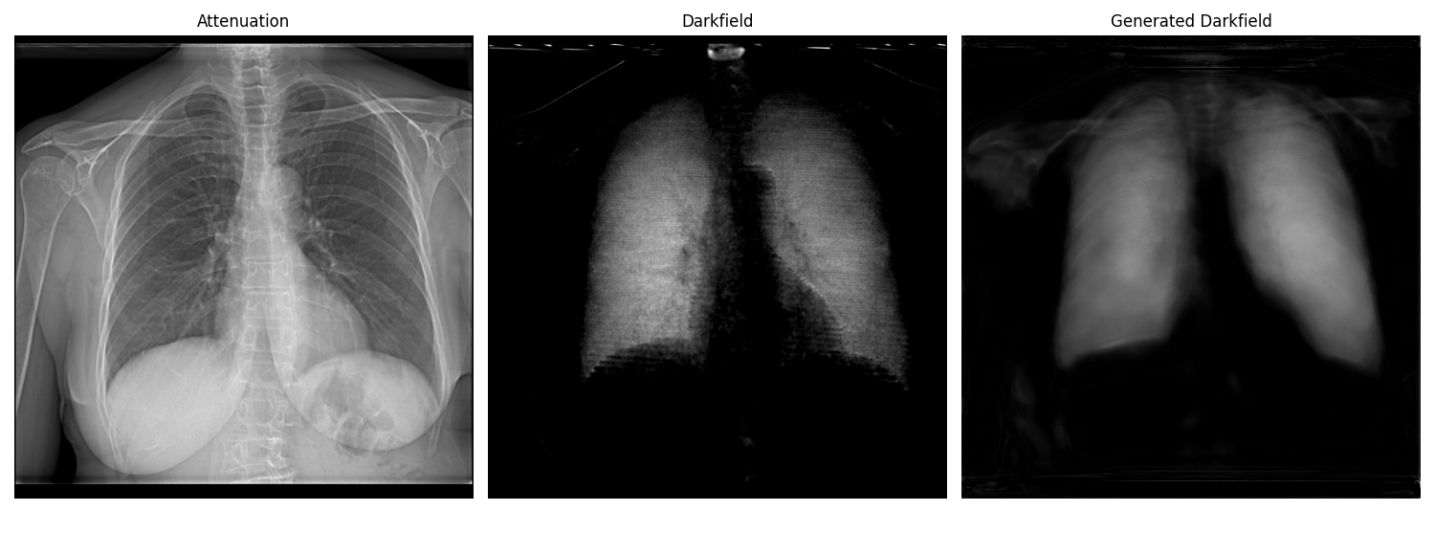
Conventional X-rays primarily highlight density variations within tissues, but X-ray Dark-Field Radiography (DFR) accesses a different contrast mechanism – the scattering of X-rays by microstructures. This technique reveals details about tissue organization at a scale invisible to standard radiography, potentially enabling earlier detection of diseases like fibrosis or even subtle changes predating anatomical alterations. However, interpreting DFR images is considerably more complex; the resulting data isn’t a direct representation of tissue structure, but rather a map of scattered X-ray intensity. Consequently, advanced computational algorithms and image processing are essential to deconvolve the scattering signal, remove artifacts, and ultimately translate the raw data into clinically meaningful information about the underlying tissue microstructure. These sophisticated techniques are actively being developed to unlock the full potential of DFR as a complementary diagnostic tool.
Deep Learning: Extracting Signal from Noise
Deep learning techniques, particularly convolutional neural networks (CNNs), are increasingly utilized in radiographic image reconstruction and interpretation due to their capacity to learn complex feature representations from data. These methods excel at identifying and enhancing subtle image features often obscured by noise or limitations of the imaging process. This is achieved through the network’s ability to learn hierarchical representations, effectively filtering noise and amplifying relevant details. Applications include improved visualization of microstructures in materials science, enhanced medical imaging for diagnostic purposes, and improved non-destructive testing capabilities by revealing previously undetectable anomalies. The power of these techniques stems from their ability to move beyond traditional image processing methods, offering a data-driven approach to feature extraction and image enhancement.
Deep learning models used for image enhancement and reconstruction require substantial amounts of labeled data to achieve optimal performance. Acquiring and annotating these datasets can be both time-consuming and expensive, often presenting a significant bottleneck in model development. To mitigate this limitation, synthetic data generation techniques are employed as a crucial data augmentation strategy. By creating artificial datasets that mimic the characteristics of real images, the volume of training data is effectively increased, enabling models to generalize more effectively and improve their ability to reconstruct or enhance images, particularly in scenarios where real-world data is scarce or difficult to obtain.
Quantitative assessment of image reconstruction accuracy relies on established metrics including Mean Squared Error (MSE), Peak Signal-to-Noise Ratio (PSNR), and Structural Similarity Index (SSIM). Lower MSE values indicate reduced reconstruction error, while higher PSNR values denote improved signal strength relative to noise. The framework detailed herein demonstrates a consistent trend of decreasing MSE and increasing PSNR across successive model stages. This progression signifies a corresponding enhancement in image fidelity, as the reconstructed images more closely approximate the ground truth data with each stage of processing.
The Structural Similarity Index (SSIM) achieved by the framework in the final reconstruction stage is 0.5. SSIM is a perceptual metric that quantifies the similarity between two images, with a value of 1.0 indicating perfect similarity and 0.0 indicating no similarity. A score of 0.5 indicates moderate structural similarity between the generated dark-field images and the real dark-field images used for comparison; while not representing a high degree of fidelity, it suggests the framework successfully captures some of the essential structural elements present in the original data. This level of similarity, combined with observed decreases in Mean Squared Error and increases in Peak Signal-to-Noise Ratio, indicates a potential for further refinement of the reconstruction process to enhance image quality.
Beyond Prediction: Quantifying the Limits of Certainty
Uncertainty Quantification (UQ) is a critical component of reliable deep learning-based image analysis because it provides a means to differentiate between genuine features within an image and spurious results caused by sensor noise, image artifacts, or limitations of the model itself. Without UQ, a deep learning system may confidently output incorrect interpretations, particularly in applications where misdiagnosis or inaccurate measurement carries significant risk. By assigning a measure of confidence to each prediction, UQ enables informed decision-making; high uncertainty flags potentially unreliable results requiring further investigation or alternative analysis, while low uncertainty indicates a robust and trustworthy outcome. This is achieved through probabilistic modeling, where the output of the deep learning system is not a single value, but rather a probability distribution reflecting the range of possible outcomes and their associated likelihoods.
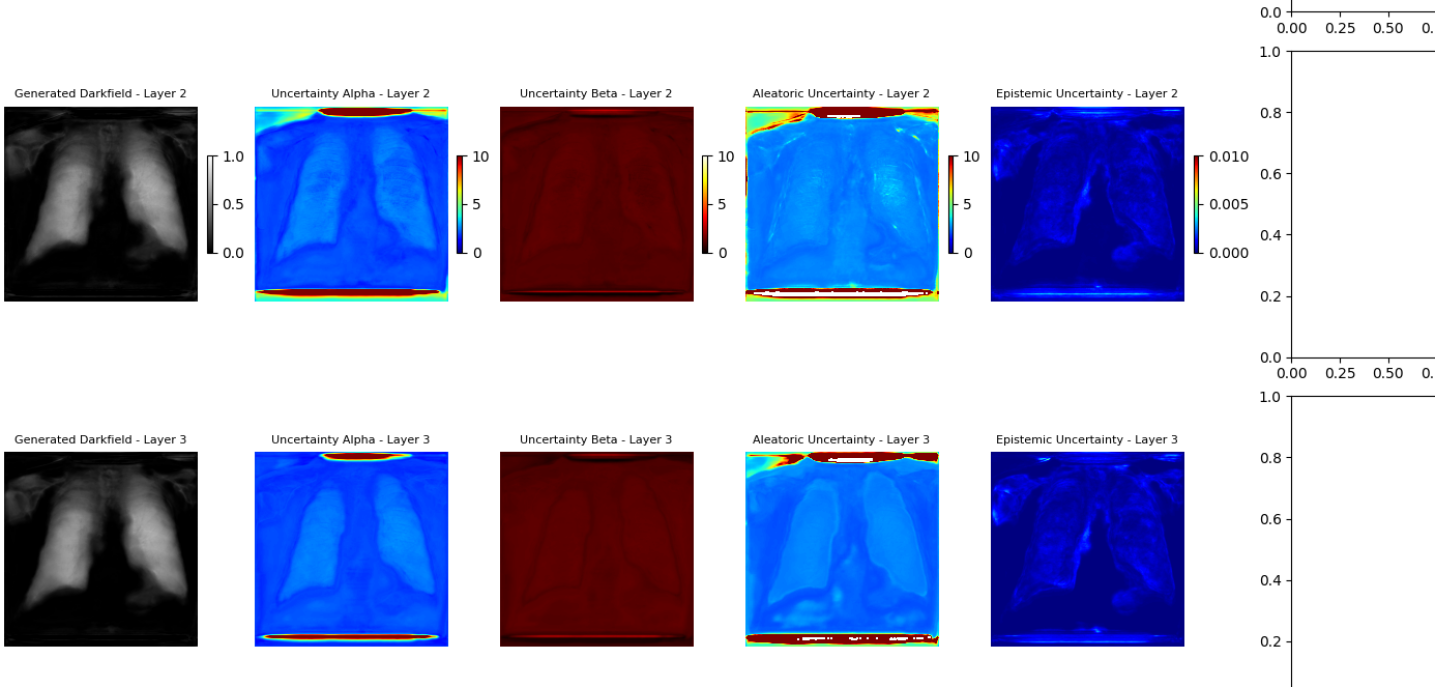
Uncertainty quantification in deep learning-based image analysis necessitates differentiating between aleatoric and epistemic uncertainty. Aleatoric uncertainty represents the inherent randomness within the data itself, encompassing noise and sensor limitations; this is irreducible even with perfect model knowledge. In contrast, epistemic uncertainty arises from a lack of knowledge within the model, resulting from limited training data or model capacity. Effectively modeling both types of uncertainty is crucial for reliable image analysis, as it allows systems to distinguish between genuine signal and uncertainty stemming from either data limitations or model inadequacies. Quantifying these uncertainties provides a more complete assessment of prediction reliability than traditional deterministic approaches.
Aleatoric uncertainty, representing inherent data noise, is effectively modeled using the Generalized Gaussian Distribution (GGD), which allows for capturing non-Gaussian error distributions commonly found in image data. The GGD is parameterized by both a mean and a shape parameter, enabling flexible adaptation to varying noise characteristics. In contrast, epistemic uncertainty, arising from a model’s lack of knowledge, is estimated via Monte Carlo Dropout. This technique involves applying dropout – randomly deactivating neurons – during both training and inference, and repeating the inference process multiple times. The variance in the resulting predictions provides a quantifiable measure of the model’s uncertainty, with higher variance indicating greater epistemic uncertainty; effectively, it simulates a Bayesian model approximation without requiring full Bayesian inference.
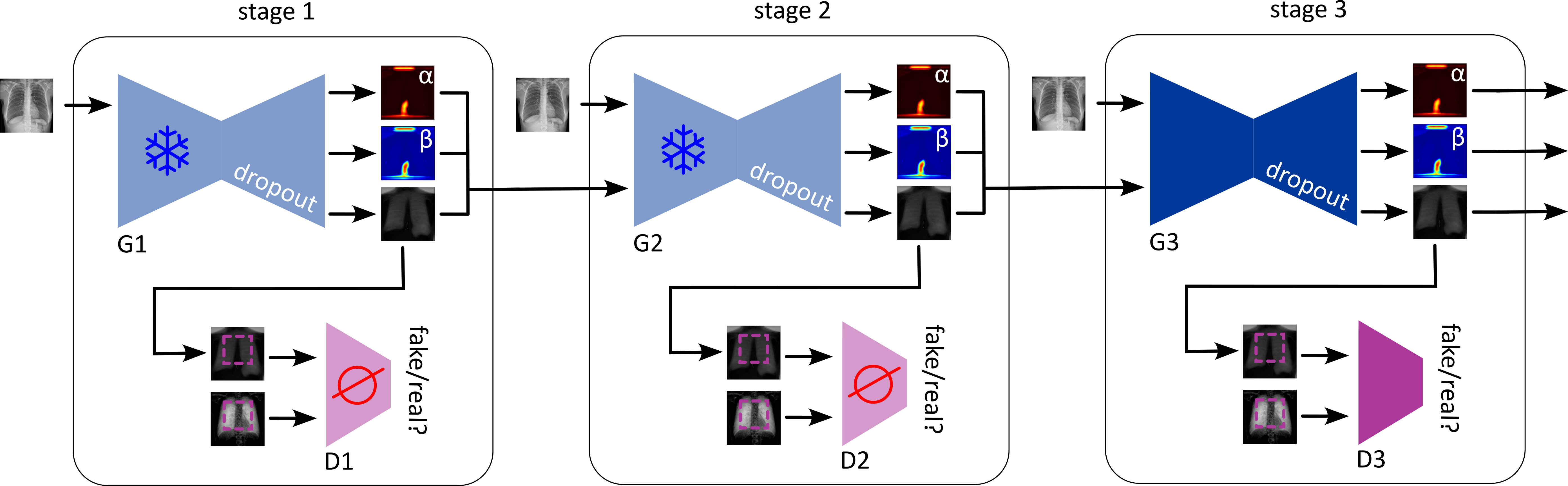
An Uncertainty-Guided Progressive GAN leverages quantified uncertainty – both aleatoric and epistemic – to iteratively refine image reconstruction. During training, the model uses uncertainty estimates as a loss function component, penalizing reconstructions with high uncertainty and guiding the generator towards more confident outputs. This is achieved through a feedback loop where the GAN’s discriminator is trained to not only distinguish real from generated images, but also to predict the uncertainty associated with each reconstruction. Higher uncertainty predictions from the discriminator result in a stronger gradient signal to the generator, prompting it to produce images that better align with the underlying data distribution and reduce overall uncertainty. This process results in improved image quality, as indicated by standard metrics like PSNR and SSIM, and a more reliable confidence score associated with each reconstructed image.
From Validation to Impact: A New Standard in Diagnostic Clarity
The true strength of this innovative framework lies in its capacity to perform effectively on data it hasn’t previously encountered. Evaluation using the National Institutes of Health (NIH) Chest X-ray Dataset – a large, independent collection of medical images – confirms the model’s robust generalization capabilities. This ability to extrapolate beyond the initial training set is critical for real-world clinical application, where imaging data invariably exhibits variations in acquisition parameters, patient demographics, and disease presentation. Successful performance on external datasets suggests the model isn’t simply memorizing training examples, but rather learning underlying patterns indicative of disease, paving the way for a more adaptable and reliable diagnostic tool.
The generative model benefits from a carefully designed training regimen employing progressive learning, a technique that steadily increases the complexity of image reconstruction. Initially, the model learns to generate low-resolution images, gradually refining its capabilities to produce high-fidelity outputs. This incremental approach not only stabilizes the training process but also significantly improves the model’s robustness against noise and variations in input data. By building upon simpler representations, the generative adversarial network avoids getting trapped in local optima and achieves a more generalized understanding of the underlying image features, ultimately leading to enhanced performance on both familiar and unseen datasets.
The pursuit of clinically relevant image reconstruction necessitates more than just structural accuracy; realistic texture and noise patterns are crucial for mirroring natural biological variations and avoiding the artificial smoothness often seen in generated images. This framework addresses this challenge through a Residual Consistency Loss, which actively encourages the preservation of subtle image details during the reconstruction process. By minimizing the difference between the residual noise in the original and reconstructed images, the system effectively learns to replicate the inherent stochasticity present in medical scans. This results in images exhibiting greater visual fidelity – appearing more natural to the trained eye of a radiologist – and crucially, improving the model’s ability to highlight subtle anomalies that might otherwise be obscured by overly-smoothed reconstructions, ultimately bolstering diagnostic confidence and potentially leading to more accurate assessments.
The culmination of this research yields a diagnostic tool poised to enhance the precision of medical imaging analysis. By generating higher-quality reconstructions – particularly in challenging scenarios with limited or noisy data – the framework minimizes the potential for misinterpretation and, consequently, diagnostic errors. This improved reliability translates directly to more informed clinical decision-making, enabling physicians to accurately identify pathologies and tailor treatment plans with greater confidence. Ultimately, the potential impact extends to improved patient outcomes, as earlier and more accurate diagnoses facilitate timely interventions and contribute to more effective healthcare delivery. The system doesn’t merely present an image; it offers a clearer, more informative representation of the patient’s condition, empowering medical professionals to provide the best possible care.
The pursuit of synthetic image generation, as demonstrated in this work, often leads to layers of complexity. However, the core strength of this framework lies in its deliberate incorporation of uncertainty – both aleatoric and epistemic. This mindful simplification aligns with a fundamental principle: clarity over complication. As Henri Poincaré stated, “It is better to know little, but to know it well.” The researchers acknowledge the inherent uncertainties within the data and model, rather than attempting to mask them, thereby producing a more robust and interpretable result. This emphasis on ‘knowing little well’ resonates deeply with the aim to distill the essential information from complex radiographic data.
Future Directions
The synthesis of dark-field radiographs from conventional images, while elegantly addressed by this work, merely shifts the locus of uncertainty. The framework’s incorporation of both aleatoric and epistemic uncertainty is not a resolution, but a formal acknowledgement of what was previously implicit. The true challenge lies not in quantifying ignorance, but in minimizing it. Subsequent research must address the fundamental gap between generated data and true physical processes-a divergence currently obscured by the metrics of image similarity.
A productive avenue involves exploring the limits of the generative model itself. The reliance on adversarial training, while effective, introduces instability and a sensitivity to hyperparameter selection. A more robust approach might consider alternative generative models-variational autoencoders, perhaps-that prioritize a faithful representation of the underlying data distribution over perceptual realism. The pursuit of ‘perfect’ image synthesis is, predictably, a distraction.
Ultimately, the value of this work will be determined not by its ability to create images, but by its contribution to improved diagnostic accuracy and reduced patient exposure. The logical extension, then, is a rigorous clinical validation-a demonstration that these synthetically augmented datasets genuinely enhance the performance of downstream tasks, and are not merely a testament to the ingenuity of the algorithm. Emotion is, after all, a side effect of structure, and the structure must be demonstrably sound.
Original article: https://arxiv.org/pdf/2601.15859.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- 39th Developer Notes: 2.5th Anniversary Update
- The 10 Most Beautiful Women in the World for 2026, According to the Golden Ratio
- TON PREDICTION. TON cryptocurrency
- Bitcoin’s Bizarre Ballet: Hyper’s $20M Gamble & Why Your Grandma Will Buy BTC (Spoiler: She Won’t)
- Gold Rate Forecast
- Lilly’s Gamble: AI, Dividends, and the Soul of Progress
- Celebs Who Fake Apologies After Getting Caught in Lies
- USD PHP PREDICTION
- Elon Musk Calls Out Microsoft Over Blizzard Dev Comments About Charlie Kirk
- VSS: A Thousand Bucks & a Quiet Hope
2026-01-23 15:32