Author: Denis Avetisyan
Researchers have developed a deep learning framework that combines time and frequency analysis of ECG signals for more accurate and reliable detection of atrial fibrillation.
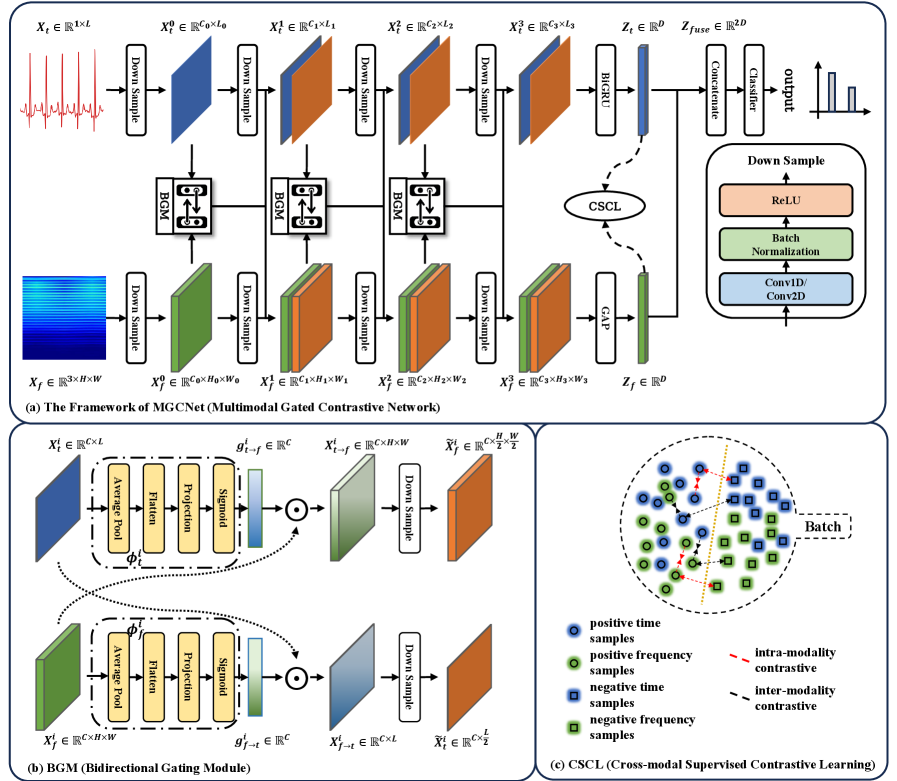
A novel deep learning architecture, MGCNet, leverages time-frequency fusion and supervised contrastive learning to improve the robustness and generalizability of atrial fibrillation detection from ECG data.
Despite advances in deep learning for cardiac arrhythmia detection, existing electrocardiogram (ECG) analysis often struggles to integrate complementary time and frequency domain information for robust and generalizable atrial fibrillation (AF) diagnosis. This work, ‘Robust and Generalizable Atrial Fibrillation Detection from ECG Using Time-Frequency Fusion and Supervised Contrastive Learning’, introduces a novel deep learning framework-MGCNet-that dynamically fuses these features via a bidirectional gating module and supervised contrastive learning. Our approach achieves improved intra-dataset robustness and strong cross-dataset generalization across multiple publicly available ECG datasets. Could this framework pave the way for more reliable and deployable AF detection systems, particularly at the point of care?
The Whispers of Chaos: Confronting the Rising Tide of Arrhythmia
Atrial fibrillation, a frequently occurring cardiac arrhythmia, now impacts over 50 million individuals globally, and its prevalence is sharply increasing in concert with the world’s aging population. This poses a significant and growing challenge to healthcare systems, demanding diagnostic solutions capable of handling a rising volume of cases. The irregular and often rapid heart rate characteristic of AF not only diminishes quality of life but also substantially elevates the risk of stroke, heart failure, and other cardiovascular complications. Consequently, the need for accurate and, crucially, scalable diagnostic tools is paramount; traditional methods are often strained by the sheer number of patients requiring monitoring, prompting research into automated systems and extended-duration recording techniques to improve detection rates and facilitate timely intervention.
Despite their established efficacy, current diagnostic standards for atrial fibrillation – namely the Standard 12-lead electrocardiogram (ECG) and Holter monitoring – present considerable practical challenges. A substantial limitation lies in the time-consuming nature of manual data review; trained personnel must meticulously analyze lengthy ECG recordings to identify subtle indicators of arrhythmia. More critically, both techniques are prone to missing paroxysmal, or intermittent, atrial fibrillation episodes. Because these irregular heartbeats can occur unpredictably and resolve spontaneously, they may not be captured during the relatively short duration of a standard ECG or even a 24-48 hour Holter monitor, leading to underdiagnosis and delayed intervention. This inherent limitation underscores the need for more sophisticated diagnostic approaches capable of continuous, automated analysis and improved detection rates for these fleeting arrhythmias.
Current diagnostic techniques for atrial fibrillation, such as standard electrocardiograms and Holter monitors, present substantial limitations in capturing intermittent or paroxysmal events, largely due to the finite recording durations and the intensive manual analysis required. This necessitates the development of automated systems capable of reliably processing extended ECG recordings-spanning days, weeks, or even months-to identify subtle patterns indicative of arrhythmia. Robust automated detection isn’t simply about increasing efficiency; it addresses a critical clinical gap by enabling more comprehensive monitoring, particularly for patients at risk of stroke who may exhibit infrequent, but dangerous, episodes of irregular heartbeat. These systems aim to alleviate the burden on healthcare professionals and improve diagnostic accuracy, ultimately facilitating timely intervention and better patient outcomes in the face of a growing prevalence of atrial fibrillation within aging populations.
The clinical significance of rapid and precise arrhythmia diagnosis extends far beyond mere identification of an irregular heartbeat; it is fundamentally linked to stroke prevention and enhanced patient well-being. Atrial fibrillation, if left undetected, dramatically increases the risk of thromboembolic events, including stroke, which can lead to significant disability or mortality. Consequently, advancements in diagnostic technologies are not simply incremental improvements, but critical necessities for mitigating this serious health threat. Earlier detection allows for timely initiation of anticoagulation therapy, effectively reducing stroke risk and improving long-term outcomes for individuals susceptible to these cardiac irregularities. The pursuit of more robust and automated diagnostic systems, therefore, represents a vital step toward proactive cardiovascular care and a demonstrably improved quality of life for a growing patient population.
Decoding the Rhythm: Deep Learning’s Ascent in ECG Analysis
Deep learning models, particularly convolutional neural networks (CNNs) and recurrent neural networks (RNNs), demonstrate efficacy in atrial fibrillation (AF) detection by autonomously learning discriminative features from raw electrocardiogram (ECG) data. Traditional AF detection relies on manual feature engineering – identifying and extracting specific waveform characteristics like P-wave absence or fibrillatory waves – followed by classification using algorithms such as support vector machines or logistic regression. Deep learning bypasses this manual step; the model learns relevant features directly from the ECG signal through multiple layers of abstraction. This automated feature extraction reduces reliance on domain expertise and can capture subtle, complex patterns indicative of AF that might be missed by traditional methods. The learned features are then used by the model for classification, predicting the presence or absence of AF with high accuracy, as demonstrated in numerous clinical studies.
Electrocardiogram (ECG) signal analysis utilizes both time domain and frequency domain approaches to characterize cardiac electrophysiology. Time domain analysis directly examines the amplitude of the ECG signal over time, identifying features such as P-waves, QRS complexes, and T-waves, and measuring their durations and amplitudes to assess cardiac function and identify arrhythmias. Conversely, frequency domain analysis, typically achieved through techniques like the Fourier Transform, decomposes the ECG signal into its constituent frequencies, revealing the spectral content and allowing for the identification of frequency-specific abnormalities or patterns that may not be readily apparent in the time domain. Each approach provides complementary information; time domain analysis offers precise morphological details, while frequency domain analysis highlights the underlying spectral characteristics of cardiac activity, improving diagnostic accuracy when used in conjunction.
One-dimensional Convolutional Neural Networks (1D-CNNs) are well-suited for direct processing of raw electrocardiogram (ECG) waveforms, treating the signal as a sequential time series and learning temporal patterns. In contrast, two-dimensional CNNs (2D-CNNs) are utilized when ECG data is transformed into a visual representation, such as a spectrogram, which maps signal frequency over time. Spectrograms allow 2D-CNNs to extract features based on spectral characteristics of cardiac events, effectively analyzing frequency components and their evolution. This approach requires an initial conversion of the 1D time-series data into a 2D image format suitable for 2D convolutional operations, enabling the network to identify patterns in the frequency domain that may not be readily apparent in the raw time-domain signal.
The Short-Time Fourier Transform (STFT) is a signal processing technique used to analyze changes in signal frequencies over time. Applied to electrocardiogram (ECG) data, STFT decomposes the ECG signal into its constituent frequencies at specific time intervals, producing a spectrogram – a visual representation of the signal’s frequency content as it evolves. This conversion from the time domain to the frequency domain allows for the identification of spectral characteristics associated with cardiac events such as P-waves, QRS complexes, and T-waves. Analyzing these spectral features can reveal subtle changes indicative of arrhythmias or other cardiac abnormalities that may not be readily apparent in the raw time-domain ECG waveform. The parameters of the STFT, including window size and overlap, directly influence the time and frequency resolution of the resulting spectrogram, necessitating careful selection based on the characteristics of the ECG signal being analyzed.
MGCNet: Forging a New Path in Robust AF Detection
MGCNet utilizes a deep learning architecture to address shortcomings in existing atrial fibrillation (AF) detection methods by integrating temporal and spectral features through a dynamic and reciprocal fusion process. Traditional approaches often treat these feature domains in isolation or with static fusion strategies, limiting their ability to capture the complex interplay of information relevant to AF identification. MGCNet overcomes this by employing a Bidirectional Gating Module (BGM) which allows for adaptive weighting and refinement of features from both temporal and spectral domains, enabling the network to prioritize the most informative characteristics for accurate AF detection. This reciprocal interaction facilitates a more comprehensive representation of the input signal, improving performance and robustness compared to methods with limited cross-domain information exchange.
The Bidirectional Gating Module (BGM) within MGCNet facilitates dynamic feature refinement through a reciprocal interaction between temporal and spectral feature streams. Specifically, the BGM consists of two gating mechanisms: one that uses temporal features to modulate spectral features, and another that uses spectral features to modulate temporal features. These gating mechanisms employ sigmoid functions to generate weights between 0 and 1, which are then applied element-wise to the respective feature maps. This weighted summation allows the network to selectively emphasize the most informative features from each domain, suppressing irrelevant or noisy signals and enabling a more nuanced representation for atrial fibrillation (AF) detection. The bidirectional nature of the gating ensures that information flows and influences processing in both temporal and spectral pathways, enhancing the model’s ability to capture complex relationships within the data.
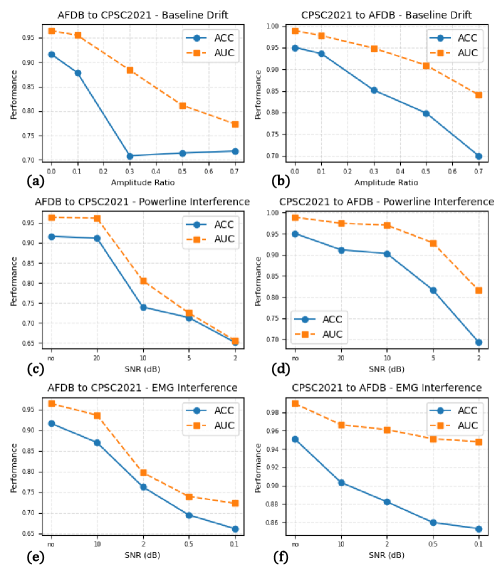
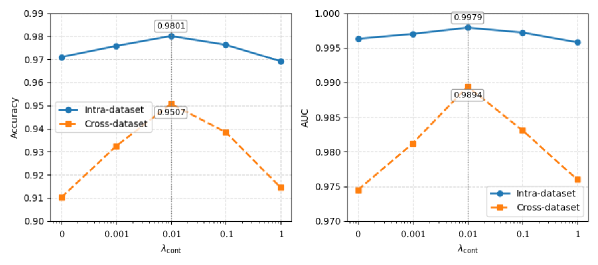
Cross-modal Supervised Contrastive Learning (CSCL) within the MGCNet architecture functions by creating a joint embedding space where temporal and spectral features are explicitly aligned. This is achieved through a contrastive loss function that encourages similar representations for corresponding features from both modalities and dissimilar representations for unrelated features. By maximizing the inter-modal similarity and intra-modal dissimilarity during training, CSCL facilitates the learning of more robust and generalizable feature representations. This approach mitigates the effects of domain shifts and improves the model’s ability to accurately detect atrial fibrillation across diverse datasets, as demonstrated by performance on the AFDB and CPSC2021 benchmarks.
Performance validation of the MGCNet architecture utilized the publicly accessible AFDB and CPSC2021 datasets. Results indicate a state-of-the-art accuracy of 0.9878 was achieved on the AFDB dataset and 0.9801 on the CPSC2021 dataset. These metrics demonstrate the model’s improved robustness and generalizability in atrial fibrillation (AF) detection across differing data distributions and acquisition protocols, exceeding the performance of previously published methods on these benchmark datasets.
Illuminating the Black Box: Visualizing MGCNet’s Reasoning
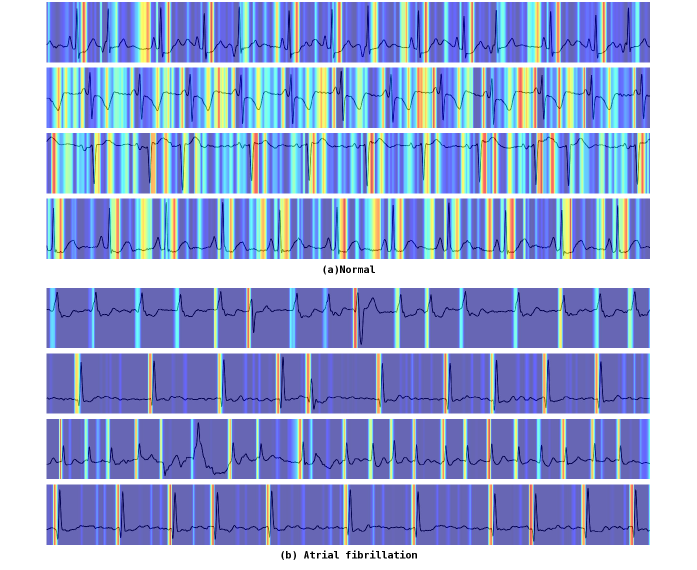
To understand how MGCNet arrives at its predictions, researchers employed Grad-CAM, a technique that generates visual heatmaps highlighting the regions of the electrocardiogram (ECG) waveform most influential to the model’s decision-making process. These visualizations effectively illuminate the specific features within the ECG that MGCNet is attending to; rather than operating as a ‘black box’, the model’s focus becomes transparent. By pinpointing these key areas, Grad-CAM not only validates the model’s learning process but also offers clinicians a crucial window into its reasoning, fostering trust and enabling informed assessments of its predictions.
Detailed visualization of MGCNet’s decision-making process demonstrates the model’s capacity to identify established hallmarks of atrial fibrillation. Through techniques like Grad-CAM, researchers observed that the network consistently focuses on specific regions of the electrocardiogram corresponding to fibrillatory waves – the chaotic electrical activity characteristic of AF – and irregular RR intervals, which represent the inconsistent timing between heartbeats. This isn’t simply pattern recognition; the model demonstrably learns and prioritizes clinically relevant features, suggesting it’s not relying on spurious correlations but instead, effectively emulating the diagnostic reasoning of experienced cardiologists. The ability to pinpoint these key indicators provides strong evidence for the model’s reliability and its potential to assist in accurate and efficient atrial fibrillation detection.
The ability to understand why a machine learning model arrives at a particular prediction is crucial for real-world application, and MGCNet’s interpretable design directly addresses this need. By visually highlighting the specific features within an electrocardiogram that drive its classifications, clinicians gain a clearer understanding of the model’s reasoning, fostering trust in its output. This transparency isn’t merely academic; it actively smooths the path for integrating MGCNet into existing clinical workflows, allowing healthcare professionals to confidently utilize its diagnostic support and ultimately enhance patient care. The resulting assurance enables a collaborative approach, where the model acts not as a black box, but as a valuable tool augmenting human expertise.
MGCNet distinguishes itself not only through its diagnostic accuracy but also through remarkable computational efficiency. The model achieves state-of-the-art performance utilizing a mere 1.96 million parameters, a significantly smaller footprint than many contemporary deep learning architectures. This compact design translates directly into a low computational complexity of 0.96 Giga Floating Point Operations (FLOPs), enabling faster processing speeds and reduced energy consumption. Such efficiency is crucial for practical deployment, particularly in resource-constrained environments or for real-time monitoring applications where timely analysis is paramount; it suggests MGCNet could be readily integrated into portable devices or scaled for high-throughput screening without substantial infrastructure requirements.
Rigorous evaluation demonstrates the exceptional diagnostic accuracy of the model, achieving an area under the receiver operating characteristic curve (AUC) of 0.9979 on the challenging CPSC2021 dataset and 0.9959 on the widely used AFDB. These metrics indicate a remarkably high capacity to correctly distinguish between normal heart rhythms and atrial fibrillation, signifying robust performance across distinct datasets and validating the model’s potential for reliable clinical application. Such high AUC values suggest the model exhibits minimal false positive and false negative rates, crucial for effective patient screening and diagnosis.
The pursuit of accurate atrial fibrillation detection, as demonstrated by MGCNet’s time-frequency fusion, feels less like engineering and more like coaxing order from inherent uncertainty. The model doesn’t find the signal, it persuades the data to reveal it. As Georg Wilhelm Friedrich Hegel observed, “The truth is the whole,” and here, the ‘whole’ necessitates integration-the melding of time and frequency domains to achieve a more complete understanding. This isn’t about eliminating noise, but embracing the totality of observation, recognizing that even apparent contradictions hold vital information. Beautiful plots can mislead, and a model performing perfectly in isolation may still crumble under the weight of real-world complexity.
What Lies Ahead?
The pursuit of automated atrial fibrillation detection, as exemplified by MGCNet, isn’t about conquering arrhythmia; it’s about building increasingly elaborate rituals to appease the inherent noise in biological signals. This work, fusing time and frequency domains, merely refines the incantation. The true challenge isn’t feature extraction, but accepting that any ‘robust’ model is simply a temporary truce with chaos. Generalizability, the holy grail, remains elusive, because data never lies; it just forgets selectively, and each new dataset represents a different forgetting.
Future efforts will likely focus on even more intricate fusion techniques, perhaps venturing into multi-modal data-incorporating everything but acknowledging that adding data doesn’t diminish uncertainty, only diffuses it. Contrastive learning, while promising, is still an act of faith-a belief that dragging similar examples closer will magically imbue the model with true understanding. The metrics-accuracy, sensitivity, specificity-are a form of self-soothing, useful for grant applications, but ultimately irrelevant in the face of the unpredictable patient.
The real frontier isn’t better algorithms, but a philosophical shift. Perhaps the goal shouldn’t be to detect atrial fibrillation, but to build systems that gracefully adapt to its presence-systems that acknowledge the limits of prediction and prioritize resilience over perfect foresight. Predictive modeling is, after all, just a way to lie to the future.
Original article: https://arxiv.org/pdf/2601.10202.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- 39th Developer Notes: 2.5th Anniversary Update
- Gold Rate Forecast
- Here’s Whats Inside the Nearly $1 Million Golden Globes Gift Bag
- The Hidden Treasure in AI Stocks: Alphabet
- TV Pilots Rejected by Networks
- The Labyrinth of JBND: Peterson’s $32M Gambit
- 20 Must-See European Movies That Will Leave You Breathless
- The Worst Black A-List Hollywood Actors
- You Should Not Let Your Kids Watch These Cartoons
- Mendon Capital’s Quiet Move on FB Financial
2026-01-18 09:12